A new method makes neural networks more streamlined, more efficient, and easier to understand—impacting research across disciplines.

Jonas Fischer, Research Group Leader on "Explainable Machine Learning" at the Max Planck Institute for Informatics. Photo: Philipp Zapf-Schramm/MPI INF
Neural networks are driving scientific progress, for example in medicine, where they help decipher genetic patterns and develop new therapeutic approaches. However, as these models grow increasingly complex, it often remains unclear why they arrive at certain predictions, posing a challenge for researchers who seek to gain new insights and validate existing knowledge. A team involving the Max Planck Institute for Informatics in Saarbrücken, Germany, has now developed a method that enhances both the efficiency and interpretability of neural networks by strategically integrating existing domain knowledge to optimize their structure.
The study, titled “Pruning Neural Network Models for Gene Regulatory Dynamics Using Data and Domain Knowledge”, was authored by Jonas Fischer, a research group leader in Explainable Machine Learning in the Computer Vision and Machine Learning department at the Max Planck Institute for Informatics, along with Inthekab Hossain and Professor John Quackenbush from the Department of Biostatistics at Harvard T.H. Chan School of Public Health, and Rebekka Burkholz, a tenure-track faculty member at the Helmholtz Center for Information Security (CISPA) in Saarbrücken and head of the Relational Machine Learning research group.
A major challenge in modern neural networks is over-parameterization: models often contain millions to billions of parameters. While such models are highly capable, their extreme complexity makes them difficult to understand, an issue seen for example in large language models like ChatGPT. In scientific applications, this lack of transparency can significantly hinder knowledge discovery. To address this, network connections can be selectively removed to streamline models, a process known as pruning.
Previous pruning methods have primarily focused on optimizing hardware efficiency. With “Domain-Aware Sparsity Heuristic” (DASH), the researchers have introduced a method that leverages pruning specifically to improve interpretability. Their approach incorporates prior domain-specific knowledge to guide the model during training, ensuring that it learns structures aligned with domain knowledge. After an initial learning phase, the model’s connections are compared with the existing knowledge, evaluated, and unnecessary connections are removed early on. To adapt to variations in data quality or uncertainty in expert knowledge, the researchers integrated a tuning parameter, allowing for dynamic balancing between expert knowledge and the structures learned during training. This iterative process continues until the model reaches an optimal balance between sparsity (i.e. a streamlined network with only the most relevant connections) and predictive performance.
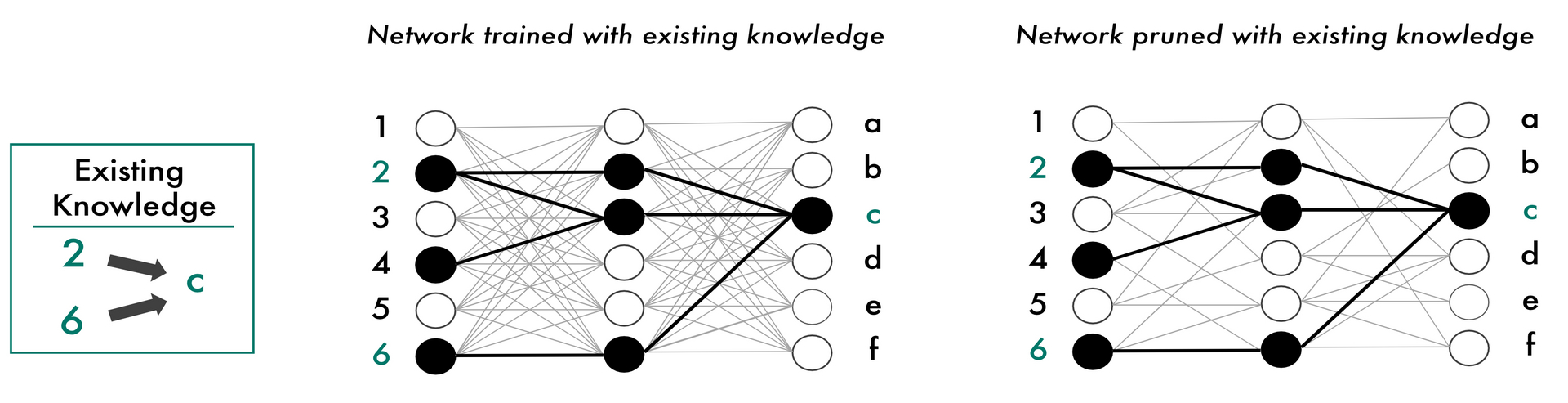
To integrate existing domain knowledge into neural networks, we can adjust the network during training so that this knowledge is reflected in the decision-making process. In the natural sciences, particularly in biology, such expert knowledge has been accumulated over decades and repeatedly validated in laboratory experiments, leading to more robust decisions. Our method now enables us to “prune” the network based on existing knowledge—removing unnecessary connections (right), making decisions more transparent and potentially enhancing the performance of neural networks. Author: Jonas Fischer
The researchers initially applied their method to gene regulation, where interpretability is particularly valuable. “Measuring gene regulation is extremely costly, but new insights in this field could be crucial for medical applications such as cancer therapies or pharmaceutical development,” explains Jonas Fischer.
A key advantage of DASH is knowledge extraction, that is, the ability to derive meaningful insights from machine-learned models. By integrating machine learning with validated expert knowledge, the model structure becomes more transparent, making it easier for researchers to analyze relationships within networks. “Our method could help uncover biologically relevant patterns that are often obscured in over-parameterized or purely efficiency-driven models,” says Fischer. The regulatory patterns encoded in the learned model can then be further analyzed and experimentally validated for their scientific significance.
In their study, the researchers tested DASH on synthetic data and on real-world datasets related to yeast cell cycle dynamics and human breast cancer, using gold-standard experimental data as a benchmark. Their results show that DASH significantly outperforms previous methods—not only in terms of the scientific validity of the resulting models but also in data efficiency and predictive performance. Notably, their model identified a strong link between the heme signaling pathway and breast cancer progression. This pathway had previously been associated withanti-tumor effects, but the DASH-pruned model revealed two additional potential regulators of the process, both of which could serve as targets for drug development. There is already an existing drug available for one of these regulators, offering a potential opportunity for drug repurposing in breast cancer treatment.
The team of researchers believes that their method can be applied beyond gene regulation and serve as a blueprint for optimizing neural networks in various scientific disciplines, particularly in natural sciences, where high-quality, well-established domain knowledge exists. Their study further demonstrates that DASH remains effective even when prior knowledge is incomplete or slightly incorrect, still delivering robust results. However, heavily flawed expert knowledge can impact the quality of the generated models, as Fischer notes: “DASH performs well even with imperfect domain knowledge, but when the input data is significantly flawed, the model’s outputs can become biased.”
The study was presented in December 2024 at the Conference on Neural Information Processing Systems (NeurIPS) in Vancouver, Canada—one of the world’s leading conferences on artificial intelligence, with a particular focus on neural networks. Additionally, earlier work by the team, which laid the foundation for this research, was highlighted by the U.S. National Cancer Institute (NCI), the leading federal agency for cancer research, as an important result in the field for the year 2024.
Original publication:
I. Hossain, J. Fischer, R. Burkholz, J. Quackenbush (2024): „Pruning neural network models for gene regulatory dynamics using data and domain knowledge“. 38th Conference on Neural Information Processing Systems (NeurIPS).
https://proceedings.neurips.cc/paper_files/paper/2024/hash/d52d2281babd36913643392a09a56832-Abstract-Conference.html
Scientific Contact:
Dr. Jonas Fischer
Max Planck Institute for Informatics
Email: jonas.fischer[at]mpi-inf.mpg.de
Editor:
Philipp Zapf-Schramm
Max Planck Institute for Informatics
Tel: +49 681 9325 5409
Email: pzs@mpi-inf.mpg.de
