EpiExplorer and RnBeads: Integrative Analysis of Epigenomic Data
Spotlight: Released Wednesday, 08 May 2013
Felipe Albrecht and Fabian Müller
Our DNA encodes how our cells behave and what they look like. However, there is a multitude of additional layers of control that determine the fate of a cell. Chemical modifications of the DNA itself (such as DNA methylation) or its molecular scaffold (such as histone modifications) regulate the ways in which the DNA is packed and processed in the nucleus of the cell, and thus are responsible for the cell's biological properties (phenotype). Two software packages developed at the Max Planck Institute for Informatics facilitate the interpretation and integration of such epigenetic layers of molecular information: EpiExplorer allows for an integrative view and interactive exploration of genomic and epigenomic features based on technology that has been developed in the context of internet search engines for searching information in a fast and intuitive fashion. RnBeads is a comprehensive software package supporting the analysis of experimentally obtained DNA methylation data.
EpiExplorer: A tool for the efficient search and comparison of genomic and epigenomic features
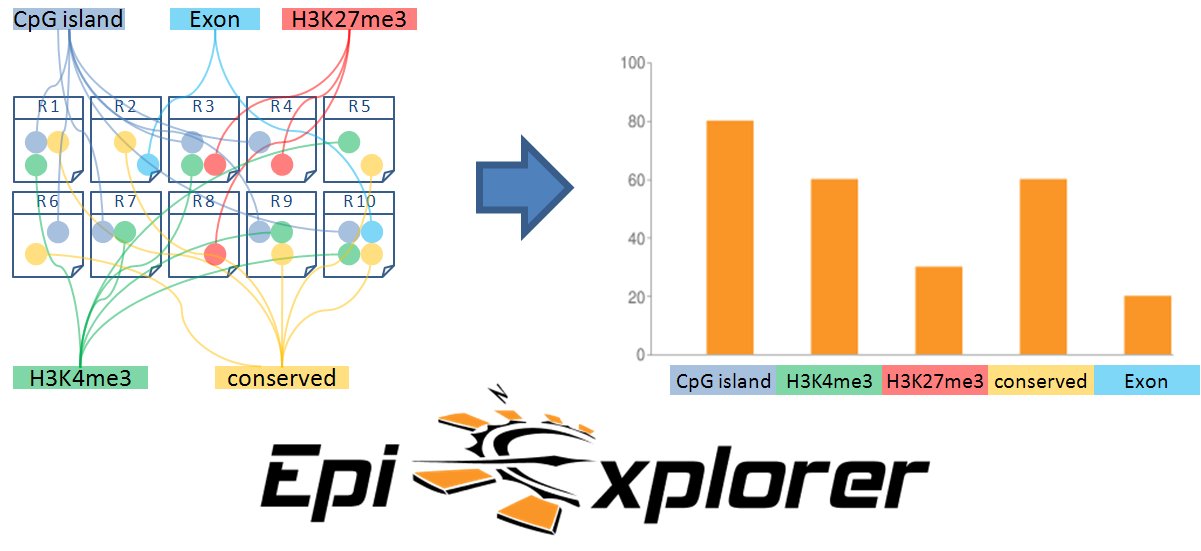
Recent technological advances are leading to an explosion in the amount of quantitative biological data available for a large number of individuals and cell types. Major bioinformatics challenges include the integration of many heterogeneous datasets comprising sequence information on the molecular architecture of the packed genome (chromatin) and on its epigenomic regulation as well as facilitating the discovery of molecular interrelations between genomic and epigenomic features. EpiExplorer is a web-based tool that enables scientists to explore a multitude of large datasets in a fast and intuitive fashion. EpiExplorer's key strength lies in supporting exploratory hypothesis generation using a broad range of genome-wide data analyses performed in real time over the Internet. The tool integrates a variety of genomic and epigenomic maps and helps scientists identify landmarks along the genome sequence characterizing regions in the genome that are of particular interest. EpiExplorer uses a versatile text indexing scheme in order to perform comprehensive analysis and visualization over the web in a matter of seconds. Datasets can also be compared quantitatively and users can incorporate their own datasets into the analysis.
Comprehensive analysis of DNA methylation data using RnBeads
DNA methylation comprises an important layer of epigenomic regulation of genomic programs and is probably the best studied of all epigenomic marks. It is a major contributor to embryonic development, and aberrant methylation patterns have been associated with complex diseases like cancer. Various experimental assays have been developed for genome-wide analysis of DNA methylation patterns. RnBeads is a software package that enables both wet lab scientists and computationally inclined scientists to run the entire analysis pipeline on data originating from DNA methylation experiments: Experimental biases can be detected and DNA methylation fingerprints can be quantified, visualized and compared. For instance, genomic regions that exhibit differential methylation fingerprints in cancerous tissue compared to normal cells can be identified and annotated with biological knowledge. Genomic regions of interest can be exported to EpiExplorer where comparison with other (epi)genomic maps is facilitated. While RnBeads' default workflow requires no expert knowledge, due to its modular design it is also highly configurable for custom analysis. The tool generates comprehensive and interpretable results in HTML format, thus facilitating easy tracking and comparison of analyses as well as exchanging results with collaboration partners.

